Abstract
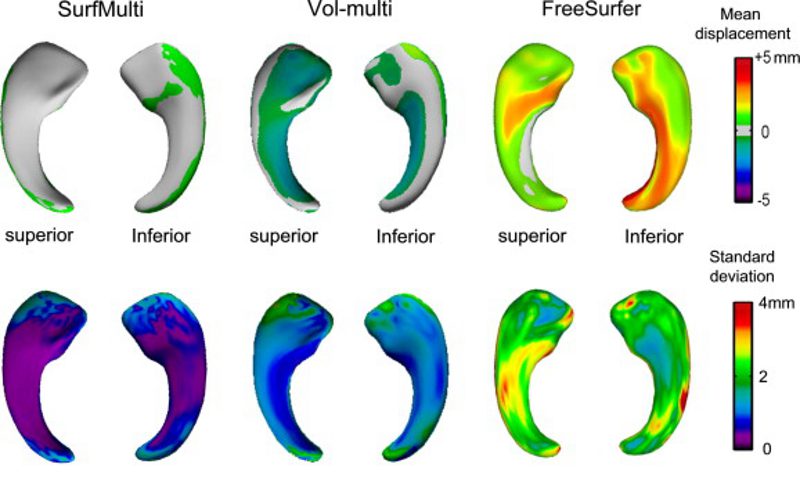
In drug-resistant temporal lobe epilepsy (TLE), detecting hippocampal atrophy on MRI is crucial as it allows defining the surgical target. In addition to atrophy, about 40% of patients present with malrotation, a developmental anomaly characterized by atypical morphologies of the hippocampus and collateral sulcus. We have recently shown that both atrophy and malrotation impact negatively the performance of volume-based techniques. Here, we propose a novel hippocampal segmentation algorithm (SurfMulti) that integrates deformable parametric surfaces, vertex-wise modeling of locoregional texture and shape, and multiple templates in a unified framework. To account for inter-subject variability, including shape variants, we used a library derived from a large database of healthy (n=80) and diseased (n=288) hippocampi. To quantify malrotation, we generated 3D models from manual hippocampal labels and automatically extracted collateral sulci. The accuracy of SurfMulti was evaluated relative to manual labeling and segmentation obtained through a single atlas-based algorithm (FreeSurfer) and a volume-based multi-template approach (Vol-multi) using the Dice similarity index and surface-based shape mapping, for which we computed vertex-wise displacement vectors between automated and manual segmentations. We then correlated segmentation accuracy with malrotation features and atrophy. SurfMulti outperformed FreeSurfer and Vol-multi, and achieved a level of accuracy in TLE patients (Dice=86.9%) virtually identical to healthy controls (Dice=87.5%). Vertex-wise shape mapping showed that SurfMulti had an excellent overlap with manual labels, with sub-millimeter precision. Its performance was not influenced by atrophy or malrotation (|r|<0.20, p>0.2), while FreeSurfer (|r|>0.35, p<0.0001) and Vol-multi (|r|>0.28, p<0.05) were hampered by both anomalies. The magnitude of atrophy detected using SurfMulti was the closest to manual volumetry (Cohen's d: manual=1.71, t=7.6; SurfMulti=1.60, t=7.0; Vol-multi=1.38, t=6.1; FreeSurfer=0.91, t=3.9). The high performance of SurfMulti regardless of cohort, atrophy and shape variants identifies this algorithm as a robust segmentation tool for hippocampal volumetry.